 |
|||||||
 |
|||||||
Specific Objectives of the Research Program 1. To generate, either from hybridoma culture or from phage display single-chain antibody libraries, a series of monoclonal antibodies that recognize diverse carbohydrate structures within plant cell walls. 2. To characterize the epitope structures recognized by antibodies that each bind to unique glycostructures within the plant cell wall. 3. To make the monoclonal antibodies available to the scientific community. Introduction All growing plant cells are surrounded by a polysaccharide-rich primary wall (O'Neill and York, 2003). The structural complexities of the wall polysaccharides suggest that plants invest significant amounts of energy and nutrient resources to synthesize and modify their walls during growth and development (Doblin et al. 2003; Rose et al. 2003). Indeed, recent estimates suggest that more than 2000 genes may encode proteins involved in these processes (Carpita et al. 2001). Many of these genes belong to multi-gene families whose members have distinct patterns of expression among plant cells and tissues, The functions of all but a handful of genes involved in cell wall synthesis and modification remain unknown (The Arabidopsis Genome Initiative, 2000; Perrin et al. 2001). Thus, functional genomics of the primary wall is a major challenge for plant biologists. The Primary Wall 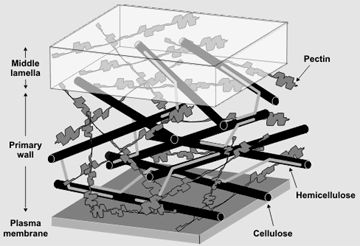 Primary walls give shape and structure to plant cells. tissues, and ultimately organs. These walls are sufficiently strong to prevent the cell from rupturing yet must be flexible and plastic to accomodate growth (Cosgrove, 2003). There is increasing evidence that primary walls have important roles in the biology of plant cells particularly in their development and differentiation. Thus, increasing our knowledge of the structure and function of the macromolecular components of primary walls and how their synthesis is co-ordinated and regulated is requiired to understand how these components interact to form a functional wall. Primary walls give shape and structure to plant cells. tissues, and ultimately organs. These walls are sufficiently strong to prevent the cell from rupturing yet must be flexible and plastic to accomodate growth (Cosgrove, 2003). There is increasing evidence that primary walls have important roles in the biology of plant cells particularly in their development and differentiation. Thus, increasing our knowledge of the structure and function of the macromolecular components of primary walls and how their synthesis is co-ordinated and regulated is requiired to understand how these components interact to form a functional wall.Models of primary walls have remained relatively unchanged since the work of Peter Albersheim and his colleagues (Keegstra et al. 1973), yet still provide a framework for the macromolecular organization of the wall (see Figure on right - adapted form McCann and Roberts, 1991). Primary walls are believed to consist of several interconnected matrices composed of polysaccharides and (glyco)proteins. Such matrices include cellulose and associated hemicelluloses (e.g. xylglucan and galactoglucomannan) and the pectic matrix composed of homogalacturonan, rhamnogalacturonan I and rhamnogalacturonan II (O'Neill and York, 2003). Some wall polysaccharide are linear polymers composed of a single type of glycosyl reside (e.g cellulose is composed of 1,4-linked β-glucosyl residues whereas homogalacturonan is composed of 1,4-linked α-galactosyluronic acid residues). Other wall polysaccharides (e.g. xyloglucan and rhamnogalacturonan II) have a regular branching pattern. In contrast, the polysaccharide backbone of rhamnogalacturonan I and the protein backbone of arabinogalactan proteins are substituted with a diverse range of arabinosyl and galactosyl-containing oligosaccharide side chains. Despite the progress made in understanding the structures of these polysaccharides and proteoglycans determining their primary structures remains a major challenge (O'Neill and York, 2003). This is due in large part to the fact that the biosynthesis of polysaccharides is, in contrast to proteins and nucleic acids, not template-driven. Thus, a polysaccharides primary sequnce cannot be inferred from genomic data. Classical genomic approaches have been used to identify and characterize some of the genes involved in wall synthesis and metabolism (Perrin et al. 2001). Database searches for amino acid domains characteristic of glycosylhydrolases, glycosyltransferases, and nucleotide-sugar interconverting enzymes have been used to identify candidate genes. Some of these genes have been cloned and functionlly expressed. Similarly, some of these genes have been mutated or their expression suppresed in an attempt to identify their function. However, the proteins encoded by these genes are often difficult to characterize. For example, glycosyltransferases are integral membrane proteins that are expressed at low levels in most cells (Sarria et al. 2001). The lack of well characterized acceptor molecules that are required by glycosyltransferases further hampers functional characterization. Moreover, chemical methods lack the sensitivity and resolution to detect changes in wall composition and structure that may occur in small numbers of cells. In addtion plants may have compensatory mechanisms to maintain wall functionality that may obscure the effect of a mutation in a particular gene. Thus there is a need for methods to detect the prodcuts of wall biosynthesis at the cellular level that may result from mutations in wall-related genes. Monoclonal Antibodeis as Tools for Cell Wall Characterization Monoclonal antibodies are specific and sensitive reagents that have been used to monitor changes in primary wall composition and organization at the cellular and sub-cellular level (Knox, 1997; Bush and McCann, 1999; Freshour et al. 2003). For example, monoclonal antibodies have been used to show that wall composition differs amongst cell types and even in the walls surrounding a single cell. Antibodies have provided evidence that within a single wall there are sub-domains containing different glyconjugates. In addition, studies with antibodies suggest that carbohydrate epitopes of glycoproteins and polysaccharides change during plant development. However, the lack of well-characterized antibodies is a major hinderance to advances in these type of studies. To date, only a small number (~20) of monoclonal antibodies have been generated that bind to plant cell wall carbohydrate structures. Only a few of these antibodies have had their binding sites (epitopes) characterized in detail (Willats and Knox, 2003). Recent progress on several fronts make this an opportune time to build a diverse antibody toolkit for functional genomics of plant cell walls: 1. Large populations of mutants in several plant species have been generated by systematic mutagensis studies. The screening of such libraries for plants with altered wall composition/structure will be greatly facilitated by the availability of a diverse library of antibody probes against wall glycostructures. 2. The characterization of antibody binding specificity is now more tractable because of improved analytical methods, the availabilty of oligosaccharide microarrays, the availability of specifc glycosylhydrolases, and improvements in the procedures for the chemical synthesis of oligosaccharides. |
| References Bush and McCann (1999). Pectic epitopes are differentially distributed in the cell walls of potato (Solanum tuberousm) tubers. Physiol. Plant 107:201-213. Carpita et al (2001). Molecular biology of the plant cell wall: searching for the genes that define structure, architecture and dynamics. Plant. Mol. Biol. 47:1-5. Cosgrove (2003). Expansion of the plant cell. In The Plant Cell Wall (JKC Rose ed), Blackwell, pp 237-263. Doblin et al (2003). Plant cell wall biosynthesis: making the bricks. In The Plant Cell Wall (JKC Rose ed) Blackwell, pp 183-222. Freshour et al (2003). Distribution of fucose-containing xyloglucans in the cell walls of the mur1 mutant of Arabidopsis thaliana. Plant Physiol. 131:1602-1612. Keegstra et al (1973). Structure of plant cell walls. III. A model of the walls of suspension-cultured sycamore cells based on the interconnections of the macromolecular components. Plant Physiol. 51: 188-197. Knox (1997). The use of antibodies to study the architecture and developmental regulation of plant cell walls. Int. Rev. Cytol. 171: 79-120. McCann and Roberts (1991). Architecture of the primary cell wall. In The Cytoskeletal Basis of Plant Growth and Form (CW Lloyd ed), Academic Press, pp. 109-129. O'Neill and York (2003). The composition and structure of primary cell walls. In The Plant Cell Wall (JKC Rose ed) Blackwell, pp. 1-54. Perrin et al (2001). Golgi enzymes that synthesize plant cell wall polysaccharides: finding and evaluating candidates in the genomic era. Plant Mol. Biol. 47: 115-130. Rose et al (2003). Cell wall disassembly. In The Plant Cell Wall (JKC Rose ed), Blackwell, pp. 264- 324. Sarria et al (2001). Characterization of a family of Arabidopsis genes related to xyloglucan fucosyltransferases. Plant Physiol. 127: 1595-1606. The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796-815. Willats and Knox (2003). Molecules in context: probes for cell wall analysis. In The Plant Cell Wall (JKC Rose ed), Blackwell, pp. 92-110. Back to Top |
|
|
| [Home] | [People] | [Background] | [Research Plan] | [Wall Antigens] | [Antibodies] | [Facilities] | [Links] |
|
|
|
|