 |
|||||||
 |
|||||||
Generating Hybridoma Cell Lines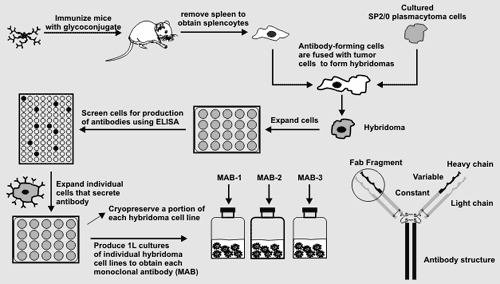 Mice will be immunized with the glycoconjugates and their sera monitored
for the presence of antibodies that recognize the glycoconjugate. When
antibody titers are sufficiently high mice splenocytes will be isolated
and then fused with SP2/0 plasmacytoma cells to form a hydridoma. Hydridoma
cells are then grown at a density of 1 cell per well. Individual antiboby-producing
cells are selected and the growth procedure repeated until antibody-producing
hydridomas each derived from a single cell are obtained. Mice will be immunized with the glycoconjugates and their sera monitored
for the presence of antibodies that recognize the glycoconjugate. When
antibody titers are sufficiently high mice splenocytes will be isolated
and then fused with SP2/0 plasmacytoma cells to form a hydridoma. Hydridoma
cells are then grown at a density of 1 cell per well. Individual antiboby-producing
cells are selected and the growth procedure repeated until antibody-producing
hydridomas each derived from a single cell are obtained.A portion of each hybridoma cell line will be cryopreseved. One litre cultures of each stable hybdridoma cell line will then be produced to obtain each monoclonal antibody. The epitope recognized by these antibodies will then be determined as will their ability to immunolabel plant cell walls. |
Screening scFv Phage-Display Libraries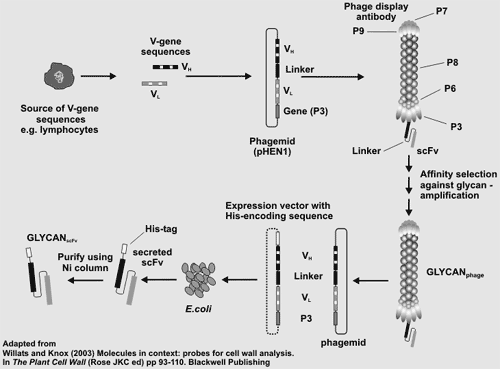 Phage-display is a method for producing recombinant monoclonal antibodies. The procedure involves the cloning of the parts of the antibody genes (VL and VH) that encode the variable domain (Fab) that is responsible for antigen recognition/binding and specifictiy. Phage-display is a method for producing recombinant monoclonal antibodies. The procedure involves the cloning of the parts of the antibody genes (VL and VH) that encode the variable domain (Fab) that is responsible for antigen recognition/binding and specifictiy. Antibody genes or gene fragments are cloned into a phage expression vector where the antibody is expressed in functional form as a fusion protein on the phage surface. Phage expressing antibodies are then selected and enriched for by their ability to bind to the antigen of interest. Phage-display libraries of antibody or antibody fragment genes provide an alternative stratergy to hybridomas for the generation of monoclonal antibodies. Phage display does not require the immunization or the sacrifice of animals and provides immediate access to the gene encoding the antibody. However, the phage themselves are inherently sticky which may lead to false positives during screening for antibody- expressing phage. Moreover, the phage library must be sufficiently large to encompass the immune repertoire of an animal to maximize the probablility of obtaing an antibody of interest. Two large libraries of single-chain antibody fragments ("Tomlinson I and J") are available from MRC geneservice. Each library contains >100 million different scFv fragments cloned in an ampicillin-resistant phagemid vector and transformed into TG1 E coli cells. Each scFv fragment is a single polypeptide comprising the VH and VL antibody domains linked by a flexible Gly-Ser hinge. We will screen these libraries for antibodies that recognize plant cell wall carbohydrate structures. Phage will be isolated after several rounds of "panning". This will ensure that at least 50% of the scFvs represented in the selected phage bind specifically to the targeted carbohydrate. The antibody-encoding gene can then be recovered and expressed to produce large amounts of the recombinant monoclonal antibody. |
Screening for Antibodies Using ELISA 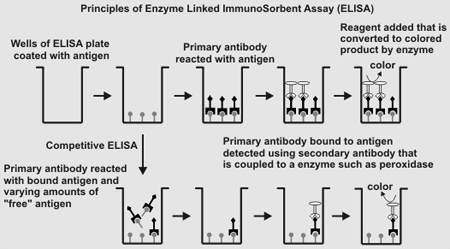 Enzyme-linked immunosorbent assay (ELISA) is a rapid and reliable method
for screening for the presence of antibodies that recognize antigens. Enzyme-linked immunosorbent assay (ELISA) is a rapid and reliable method
for screening for the presence of antibodies that recognize antigens.The basic method involves: 1. Binding the antigen to the surface of the wells of a ELISA plate. 2. Blocking any non-specific antibody binding sites with a generic protein (BSA. ovalbumin, gamma-globulin, skimmed milk). 3. Adding primary antibody solutiond to each well to allow specific antigen-antibody recognition/binding to occur. 4. Removing unbound primary antibody. 5. Adding a secondary antibody (e.g. goat anti-mouse anti IgG that is covalently linked to a protein such as horse radish peroxidase or alkaline phosphatase). This antibody binds to the primary antigen that remained bound to antigen after washing the wells. 6. Remove unbound secondary antibody 7. Determine the amount of secondary antibody bound to the wells by adding a chemical reagent that is converted by the peroxidase or phosphatase to a colored product whose absorbance is measured using a ELISA plate reader. Antibody binding specifity can be determined using a competitive ELISA in which step 3 in the above procedure is modified as follows: Primary antibody samples are pre-incubated with varying amounts of a competitor (mono-, oligo-, or polysaccharide) prior to addition to the wells of the ELISA plate that have been coated with antigen. The remainder of the ELISA is carried out as described above (steps 4 - 7). A competitor that efficiently binds to the primary antibody will reduce the amount of primary antibody that is available to bind to the immobilized antigen in the well. Quantitative comparisons of the inhibition curves provides information about which competitor is bound most efficiently by the primary antibody, and hence which competitor has the structure recognized by the antibody. |
| Polysaccharide Isolation Fragmentation of Polysaccharides Coupling of Glycans to Proteins |
Generating Hybridoma Cell Lines scFv Phage-Display Libraries ELISA |
Characterizing Antibodies Characterizing Epitope Structures |
|
|
| [Home] | [People] | [Background] | [Research Plan] | [Wall Antigens] | [Antibodies] | [Facilities] | [Links] |
|
|
| A National Science Foundation-funded (Grant No DBI-0421683) research project at The Complex Carbohydrate Research Center of The University of Georgia |