 |
 |
 Debra Mohnen |
|
 Maor Bar-Peled |
||
| Background |
| Pectic and hemicellulosic polysaccharides are assembled in the Golgi apparatus
by glycosyl transferases that catalyze the transfer of a glycosyl residue
from a nucleotide sugar to a acceptor molecule. These polysaccharides are
transported in vesicles that fuse with the plasma membrane and then released
and integrated into the pre-existing wall. In contrast, cellulose is synthesized
by protein complexes located in the plasma membrane. Cell wall polysaccharides
may be modified as the cell grows and develops. Wall biosynthesis research at the CCRC is directed towards understanding at a molecular level the factors that control the biosynthesis of pectins and hemicelluloses. These studies involve: |
|
1. Characterization of the galacturonosyl transferase(s) involved in the
synthesis of homogalacturonan |
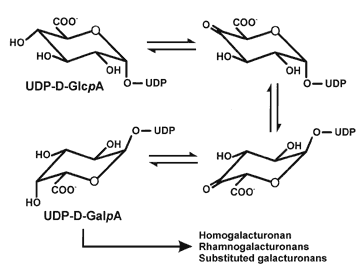
| The biosynthesis of homogalacturonan We have isolated a Golgi fraction that is enriched in a enzyme (GalAT) that transfers GalA from UDP-GalA to endogenous homogalaturonan or to exogenous oligogalactuornides. In our early studies we used 14C-labeled UDP-GalA that had been generated from 14C UDP-GlcA using a UDP-GalA 4-epimerase prepared from radish roots. We have now developed assays for this enzyme using unlabeled UDP-GalA and fluorescent labeled oligogalacturonides. The products of the reaction are monitored using polyacrylamide gel elecrophoresis (PAGE). The products are then characterized using exo and endopolygalacturonases together with size-exclusion and anion exchange chromatographies. We have shown that the catalytic site of GalAT and pectin methyl transferase are located in the lumen of the Golgi. |
 |
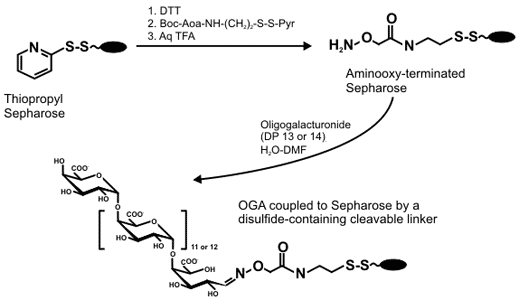
Solid-phase enzymatic synthesis of pectic oligogalacturonides Solid-phase chemistry has had an increasingly important role in the synthesis of oligosaccharides because it is readily automated and simplifies product isolation. We have combined solid-phase techniques and a Arabidopsis pectic galacturonosyltransferase to investigate chemi-enzymatic synthesis of oligogalacturonides (Guillaumie et al 2003). We have generated a Sepharose 6B gel that contains a disulfide bridged linker with a aminooxy terminal group. OGAs were then immobilized on this gel through their reducing end via oxime formation. |
 |
(GalA)14-derivatized gel was reacted with UDP-GalA in the presence of a partially purified Arabidopsis GalAT. The gels were then treated with dithiothreitol, pH 8, to reductively cleave the disulphide linkage and release thiolated OGAs. The released products were then analyzed by MALDI-TOF MS in the negative ion mode. |
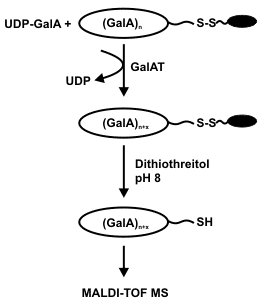 |
In the presence of limiting amounts of UDP-GalA the coupled 14mer was extended by a single GalA residue. By contrast, the coupled 13mer was extended by up to 3 GalA residues in the presence of a 5-fold excess of UDP-GalA. This demonstrates that the Arabidopsis GalAT had non-processively transferred GalA units onto the non-reducing end of the coupled OGA. |
Functional characterization of an Arabidopsis homogalacturonan galacturonosyltransferase We used peptide sequences obtained from Arabidopsis proteins enriched for homogalacturonan α-1,4-GalAT activity to identify a galacturonosyltransferase (GAUT1, At3g61130) involved in pectin biosynthesis (Sterling et al 2006). GAUT1 was shown to encode a protein with GalAT activity using human HEK293 cells that had been transiently transfected with a cDNA construct containing a N-terminally truncated version of At3g61130. The growth media from the HEK293 cells contained a protein that catalyzed the transfer of 14C-GalA from UDP-14C-GalA to exogenous oligogalacturonides. Twenty five genes encoding proteins with high similarity to GAUT1 were identified by BLAST analysis of the Arabidopsis genome (see figure below).
|
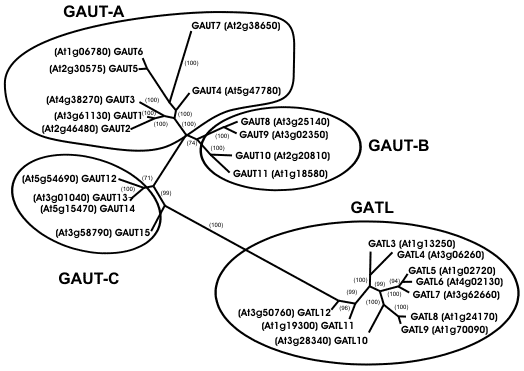 |
The GAUT genes are predicted to encode proteins with molecular masses between 61 and 78 KDa. Most of the GAUT proteins are likely to be type II membrane proteins with a transmembrane domain in their hypervariable N-terminal region. GAUT3,4, and 5 contain a N-terminal signal peptide rather than a transmembrane domain. By contrast, GAUT2 contains no N-terminal transmembrane domain and no signal peptide. The GATL genes encode proteins with molecular masses between 39 and 44 KDa and are predicted to contain only a signal peptide at their N termini suggesting they are processed into the secretory pathway. The 25 proteins in the GAUT1-related superfamily are members of the CAZy glycosyltransferase family 8. The Arabidopsis GAUT-1-related gene family consists of four clades (GAUT-A, GAUT-B, GAUT-C, and GATL, see above figure) that are clearly distinct from the other Arabidopsis glycosyltransferase family 8 proteins. Thus, we believe that the Arabidopsis family-8 proteins should be subdivided to reflect these differences (see Sterling et al 2006 for details). GAUT genes were also identified in other species including soybean (TC207293), tomato (TC154813), chickpea (CAB81547), barrel medic (TC108344), maize (TC272016), rice (Q652K2, Q5Z7Z8, BAC06990), and the moss Physcomitrella (contig3305). This data suggests that GAUT1 and other members of the GAUT1-related superfamily are conserved in vascular and avascular plants. The identification of GAUT1 as a GalAT and of the GAUT1-related gene family provides the tools required to study pectin biosynthesis at a biochemical and molecular level. |
|
A model for pectin biosynthesis |
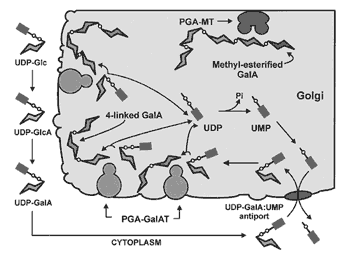 |
|
Some of the newly added 4-linked GalpA residues are methyl esterifed by
Golgi-localized pectin methyl transferases. Current Research: Functionally characterization of the GAUT family of genes. |
|
The biosynthesis of UDP-GalA UDP-GlcA 4-epimerase (UGlcAE) catalyzes the conversion of UDP-GlcA to UDP-GalA: |
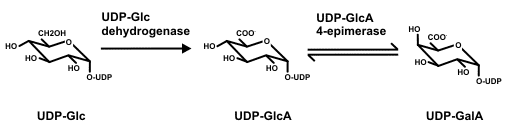 |
| A gene encoding Arabidopsis UGlcAE was identified and the recombinant enzyme biochemically characterized (Gu and Bar-Peled, 2004). The gene belongs to a family composed of six isoforms. All members of the UGlcAE family are likely to encode type-II membrane proteins and have two domains: a large conserved C-terminal catalytic region containing approximately 300 amino acids and a variable N-terminal region (~120 amino acids) containing the cytostolic, transmembrane, and stems domains. |
| The biosynthesis of UDP-D-xylose UDP-xylose is required for the synthesis of xyloglucan and xylan. UDP-D-Xylp
is itself converted to UDP-L-Araf by the combined action of a 4-epimerase
and a mutase. Thus, UDP-Xyl is likely to be one of the key nucleotide sugars
involved in the formation of both pectins and hemicelluloses. |
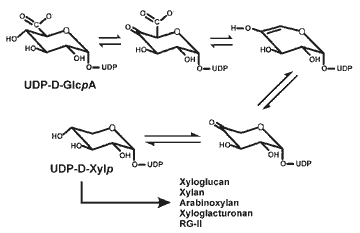 |
|
The biosynthesis of UDP-L-rhamnose |
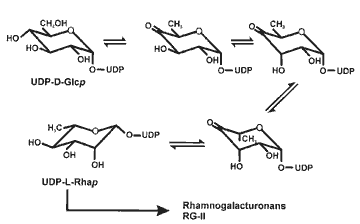 |
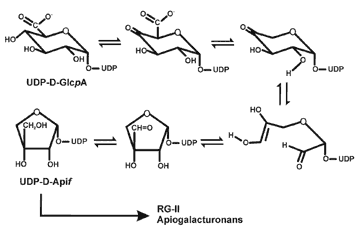
| The biosynthesis of UDP-D-Apiose Apiose is a quantitatively minor component of the primary walls of most higher plants. Nevertheless, this branched glycose is a important component of RG-II. RG-II exists in the wall as a dimer that is cross linked by a 1:2 borate-diol ester. The borate ester is formed between a apiosyl residue in each RG-II monomer. A reduction in the extent of borate cross-linking of RG-II has been shown to result in a dramatic reduction in plant growth. [Further details on RG-II are located at RG-II]. Kindel and Grisebachs research showed that UDP-D-Apif is formed from UDP-GlcA by a series of reactions that are not fully understood. However, there have been no reports of the isolation of apiosyl transferases. Current research: Generate UDP-apiose in amounts sufficient for assays to identify apiosyl transferases and their corresponding genes. |
 |
Selected references. Sterling et al (2005) Development of a filter assay for measuring homogalacturonan:α-(1,4)-Galacturonosyltransferase activity. Anal. Biochem., 343, 231-236. Sterling et al (2006) Functional identification of an Arabidopsis pectin biosynthetic homogalaturonan galacturonosyltransferase. Proc Nat Acad Sci (USA), 103, 5236-5241. Bacic (2006) Breaking an impasse in pectin biosynthesis. Proc Natl Acad Sci (USA), 103, 5639-5640. |