 |
 |
| Background Numerous defense mechanisms have evolved in plants to prevent infection by pathogenic fungi. Many of these fungi facilitate their colonization of plant tissue by producing glycanases (e.g. galacturonases, xylanses and glucanses) that fragment plant cell wall polysaccharides. The oligosaccharides that are generated by these glycanses provide the fungus with a carbon source but are also percieved by and elicit a plants defense responses. Plants produce proteins that inhibit fungal glycanses and thereby increase the life time of the biologically active oligosaccharides. The fragmentation of the fungal cell wall by plant-derived endo 1,3 β-glucanses also generates oligosaccharides that induce plant defense responses. The fungus itself may produce glucanase inhibitor proteins to prevent the degradation of its own cell wall and limit its perception by the plant. Thus, the interplay between fungal and plant glycanases and their respective inhibitors may in large part determine the outcome of attempted pathogenesis. |
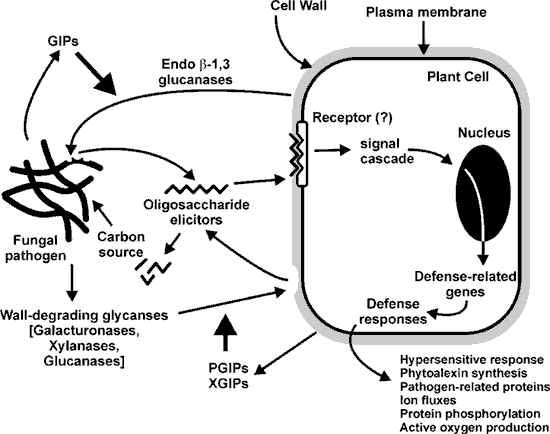 |
| Fungal endopolygalacturonases and plant polygalacturonase inhibitor proteins Pectins are a major component of the primary walls of dicots and non-graminaceous monocot cells. Pectin-degrading enzymes, including endopolygalacturonases (EPGs), are among the first glycanases to be secreted during fungal infection. EPG fragmentation of homogalacturonan (HGA) results in the transient formation of elicitor-active oligogalacturonides (OGAs) with degrees of polymerization between 9 and 15. These OGAs are rapidly converted to smaller, biologically inactive fragments by the EPGs. Thus, factors that limit fungal EPG action are likely to increase the life times and concentrations of biologically active OGAs and may result in enhanced or prolonged plant defense repsonses. Polygalacturonase inhibiting proteins (PGIPs) are leucine-rich repeat glycoproteins that are present in the apoplast of dicots and some monocots. PGIPs form reversible high-affinity complexes with fungal EPGs. The rate of hydrolysis of HGA by the PGIP-EPG complex is 1 to 2 orders of magnitude slower than the unbound enzyme. PGIPs from different plants show considerable sequence homolgy but often differ in their abilities to inhibit fungal EPGs. Several specific amino acids of PGIPs have been proposed to be involved in the binding to EPGs. However, the mechanism of the PGIP-EPG interaction is not fully understood. |
| The interaction of Aspergillus niger EPG-II with its substrate and with bean PGIP are being investigated using
surface H/2H exchange in combination with mass spectrometry. This technique provides information on which amino acids in a protein have amide groups that rapidly exchange H for 2H and provides insight into the conformational changes that occur during
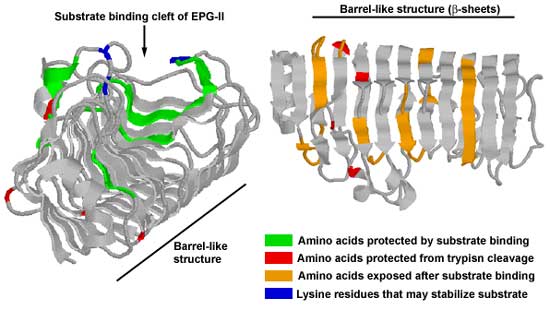
enzyme-substrate and protein-protein interactions. Our data suggests that: 1. The substrate contacts the surface of EPG-II along the cleft of a barrel-like structure previously identified by X-ray crystallography. The "barrel" spreads apart and flexes like a spring which may facilitate the hydrolysis of the glycosidic bond. A conformational change also occurs in the β-pleated sheets of the enzyme and in a portion of the α-helix that overlays the substrate bind cleft. A comparison of the fluorescence spectrum of EPG-II in the presenc or absence of substrate also indicates that binding of the enzyme to HGA results in a more structured α-helix. |
 |
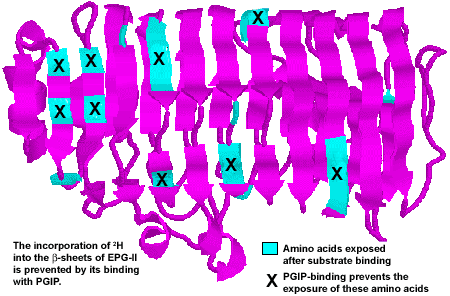
| 2. The results of 2H exchange suggest that bean PGIP contacts EPG-II on the underside of the barrel at the N-terminal side in the same region as the opened β-pleated sheet. PGIP binds to EPG-II on the side opposite the substrate binding site and may inhibit the conformational changes induced by binding of EPG-II with its substrate. The fluorescence intensity of the EPG-PGIP complex and EPG-II alone are comparable, providing further evidence that bean PGIP prevents the conformational changes that normally occur when the enzyme binds to its substrate. |
 |
| 3. We have found that under certain conditions some PGIPs actually activate
fungal EPGs and increase the rate at which they fragment HGA. For example,
EPGs A and B, the constitutive EPGs of A. niger, are activated by up to 30-fold at pH 5 by the two quantitatively major bean PGIPs. Both enzymes are fully inhibited by the PGIPs at pH 4.2. The pH-dependence of this effect may be due to ionizable groups on either protein or to the ionization state of the substrate. 4. We have obtained detailed information about the structures and locations of the high-mannose N-linked glycans that are attached to A. niger EPG-I and II. Studies with glycosylation mutants of fungal pectin methylesterases (PMEs) suggest that the N-linked sugars have little if any effect on enzyme activity. 4. We have investigated the mode of action of A. niger PME using methyl esterified oligogalacturonides and tanden MS. Our data show that methyl groups located on the internal galacturonosyl residues are the first to be hydrolyzed. Hydrolysis of the methyl group on the reducing GalA occurs only after the internal residues have been deesterified. We did not observe de-methylation of the non-reducing GalA residue. |
| Current research 1. The interaction of EPGs from Botrytis cinerea with PGIPs from bean, tomato, and pear. 2. Mechanistic studies of pectin fragmentation by various fungal EPGs. 3. The properties of PGIPs synthesized by members of the Gramineae. |
| Soybean endo-1,3 β-glucanases and Phytophora sojae glucan inhibitor proteins Albersheim and Valent (1974) observed that the fungal phytopahtogen Colletotrichum lindemuthianum secretes a protein that inhibits some of the endoglucanase secreted by the host plant. Thus, we hypothesized that fungal endo-β-1,3-glucanses inhibitor proteins (GIPs) function to protect mycelial walls from plant endoglucanses (EGases) and may also limit the production of fungal wall-derived oligosaccharides that induce the plants defense reposnses. We have used the soybean pathogen Phytophthora sojae f sp, glycines (psg) as a model system to study fungal GIPs and plant endoglucanses. A GIP (GIP1) was purified from psg culture filtrates and shown to inhibit soybean EGaseA but not EGaseB. The psg GIP did not inhibit a tobacco EGase nor did it inhibit its own EGases. We have used a genetic approach to investigate GIP genes and a biochemical approach to characterize the interaction of GIP with EGases. GIPs are a family of non-functional serine protease orthologs. Using the entire GIP1 sequence two homologs (GIP2 and GIP3) were identified. A screen of a psg BAC library suggested that there are approximately 3 - 4 GIP genes. Multiple isoforms of GIP are expressed in vitro and in infected soybean tissues. GIPs are also expressed constitutively in the walls of the fungus but no spatial distribution was apparent. GIP1 forms a compex with EGaseA in vitro and in soybean tissues. This interaction reduces the ability of the EGaseA to release from the fungal cell wall oligosaccharides that induce the plants defense responses. EGaseA and EGaseB correspond to two previously identified soybean genes. EGaseA corresponds to an ethylene regulated EGase whereas EGaseB corresponds to SGlu5. Little is known about SGlu5 expression , other than detection of mRNA in leaves, flowers, and hypocotyls. |
| Current Research 1. Alter the expression of GIPs in Phytopthora. We will use P. infestans and tomato since this fungus is easy to transform and cDNA libraries are available. 2. Spatial localization, using monoclonal antibodies, of GIPs and EGases during the infection process. 3. Determine the mechanism whereby GIP1 inhibits EGaseA. 4. Use a combination of surface plasmon resonance and the yeast two-hydrid system to determine whether the GIP1-EGaseA interaction is mediated by the "kringle" domains present in GIPs. |
| The role of endoxylanases secreted by Magnaporthe grisea (a fungal pathogen of rice) Rice blast disease (see figure on the right) is caused by the fungus Magnaporthe grisea (formerly known as Pyricularia grisea). This disease is estimated to cause losses of over $100 million annually in Asia and other rice growing areas of the world including the USA. Most infections occur on the leaves and cause necrotic lesions and tissue death or on the panicles which turn white and die before being filled with grain. M. grisea secretes several isoforms of endo-β-1,4-xylanases. We hypothesized that these enzymes are pathogenicity factors and are using a genetic approach to investigate the role of the xylanases in the infection process. We have purified and cloned two xylanases (Xyl1 and Xyl2) from M. grisea. However, deleting these genes did not abolish endoxylanase activity. We have now identifed at least 17 putative xylanses in the genome of the fungus. Six of the xylanases (Xyl 1 - 6) have been partially characterized. Three (Xyl1, Xyl3, and Xyl4) encode class 11 endo-β-xylanases while Xyl2, Xyl5, and Xyl6 encode class 10 endo-β-glucanses. Gene knock-out studies suggest that Xyl3, Xyl4, and Xyl5 are pathogenecity factors while Xyl2 may have a role in initating the host plants defense repsonses |
 |
| Current Research 1. Characterize the effects of the xylanase mutants on blast-susceptible and resistant rice cultivars 2. Over-express the xylanases in an heterologous system 3. Determine the substrate specificities of the xylanases 4. Determine whether Xyl2 or its oligoarabinoxylan products elicit defense response in rice plants 5. Determine whether rice plants synthesize xylanase-inhibitor proteins. |
|
Selected References Albersheim and Valent (1974) Host-pathogen interactions VII. Plant pathogens secrete proteins which inhibit enzymes secreted by the host capable of attacking the pathogen. Plant Physiol., 53, 684-687. Ham et al (1997) Fungal pathogens secrete an inhibitor protein that distinguishes isoforms of plant pathogenesis-related endo-β-1,3-glucanases. Plant J., 11, 169-179. Wu et al (1997) Deletion of two endo-β-1,4-xylanase genes reveals additional isozymes secreted by the rice blast fungus. Mol Plant Microbe Interact., 10, 700-708 Cook et al (1999) Fungal polygalacturonases exhibit different substrate degradation patterns and differ in their susceptibilities to polygalacturonase-inhibiting proteins. Mol Plant Microbe Interact. 12, 703-711 Stotz et al (2000) Identification of target amino acids that affect interactions of fungal polygalacturonases and thier inhibitors. Physiol Mol Plant Pathol., 56, 117-130. Rose et al (2002) Molecular cloning and characterization of glucanase inhibitor proteins. Coevolution of a counter-defense mechanism by plant pathogens. Plant Cell, 14, 1329-1345. King et al (2002) Studying protein-carbohydrate interactions by amide hydrogen/deuterium exchange mass spectrometry. Rapid Commun. Mass Spectrom. 16, 1569-1574. |