 |
 |
| Hemicelluloses | ||||
 Willie York |
|
 Stefan Eberhard |
|
|
|
Xyloglucan - composition and glycosyl sequence |
| Xyloglucan is the quantitatively predominant hemicellulosic polysaccharide
in the primary walls of dicots and non-graminaceous monocots. Xyloglucan
may account for up to 20% of the dry weight of the primary wall. Xyloglucan has a backbone composed of 1,4-linked β-D-Glcp residues. Up to 75% of thes residues are substituted at O6 with mono-, di-, or triglycosyl side chains. |
| A single letter nomenclature is used to simplify the naming of xyloglucan
side chain structures. For example, a capital G represents a unbranched Glcp residue. A capital F represents a Glcp residue that is substituted with a fucose-containing trisaccharide. The nomenclature is shown in the figure below: |
 |
| A interactive program containing xyloglucan subunit structures can be accessed by clicking here. You can enter any sequence of subunit structures (X, L, F, G etc.) and the corresponding xyloglucan structure will be shown. |
| Xyloglucans are classified as XXXG-type of XXGG-type. XXXG have three consecutive backbone residues that are subsituted with Xylp and a fourth unbranched backbone residue. XXGG xyloglucans have two consecutive branched backbone residues and two unbranched backbone residues. The glycosidic bond of the unbranched Glcp residues in XXXG and XXGG-type xyloglucans are cleaved by many endo-1,4-β-glucanases. Endoglucanase treatment of XXXG-type xyloglucans typically generates a series of well-defined oligosaccharides that have a tetrasaccharide backbone. In contrast, the types of oligosaccharide generated from XXGG-type xyloglucans depends on the substrate specificty of the enzyme and on the presence or absence of O-acetyl groups on the unbranched Glcp residues. |
| The primary walls of a wide range of dicots, non-graminaceous monocots,
and gymnosperms contain fucosylated xyloglucans with a XXXG-type structure.
The typical structures of the subunits in these xyloglucans are shown below: |
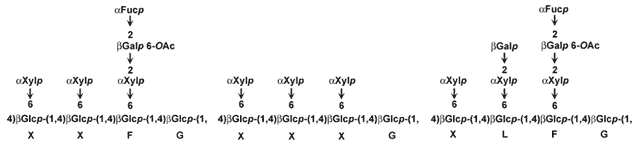 |
| Solanaceous xyloglucans together with the XXXG-type xyloglucans synthesized by members of the Olaeceae have arabinose-containing side chains but contain little if any fucose. Some structures of the subunits of the solanaceous xyloglucans are shown below: |
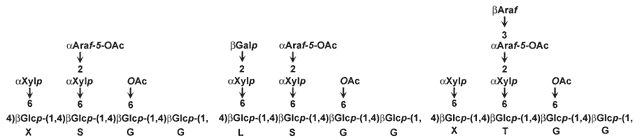 |
| We have developed a searchable 1H NMR database to facilitate the rapid identification of endoglucanase-generated xyloglucan subunit structures. |
Xyloglucan structure varies between plant species The structures of xyloglucans from several plants in the subclass Asteridae were characterized by NMR spectroscopy and mass spectrometry to determine how their structures vary in different taxonomic orders (Hoffman et al 2005). All campanulids examined, including Lactuca sativa (lettuce, order Asterales), Tenacetum ptarmiciflorum (dusty miller, order Asterales), and Daucus carota (carrot, order Apiales), produce xyloglucans that have an XXXG-type branching pattern and contain α-D-Xylp-, β-D-Galp-(1→2)-α-D-Xylp-, and α-L-Fucp-(1→2)-β-D-Galp-(1→2)-α-D-Xylp- side chains. Members of the lamiids produce atypical xyloglucans. For example, Capsicum annum (pepper) and Lycopersicon esculentum (tomato), two species in the order Solanales, and Olea europaea (olive, order Lamiales) synthesize xyloglucans that contain arabinosyl and galactosyl residues, but lack fucosyl residues. The XXGG-type xyloglucans produced by Solanaceous species are less branched than the XXXG-type xyloglucan produced by Olea europaea. We have shown that Ipomoea pupurea (morning glory, order Solanales), Ocimum basilicum (basil, order Lamiales), and Plantago major (plantain, order Lamiales) all produce xyloglucans that lack fucosyl residues and have an unusual XXGGG-type branching pattern. Furthermore, Neruim oleander (order Gentianales) produces an XXXG-type xyloglucan that contains arabinosyl, galactosyl, and fucosyl residues. The fucosylated XXXG-type xyloglucan structure is present in diverse plants, suggesting that the presence of fucosyl residues gives the plant a selective advantage. Nevertheless, laboratory grown A. thaliana mutants that do not add the fucosyl residues to XyG have no visible phenotype. Several species of plants produce xyloglucans that lack fucose, but they appear to compensate by varying other structural features. Xyloglucans synthesized by all plants examined to date have at least one of the following structural features: 1. Side chains terminated by fucosyl residues (all Gymnosperms, Rosids, and Campanulid Asterids examined) 2. Side chains terminated by arabinosyl residues (Lamiid species in the Convolvulaceae, Solanaceae, and Oleaceae families) 3. A XXGGG-type repeating core consists of a cellopentaose backbone with two side chains (Lamiid species in the Convovulaceae, Lamiaceae and Veronicaceae families). Two of these structural features (1 and 2) occur together in Nerium oleander. The combination of diverse structural features in oleander xyloglucan suggests that this species may bridge an evolutionary gap between Lamiids and plants with more typical XyG structures. This hypothesis is consistent with recent analyses indicating that divergence of the Gentianales occurred earlier than other Lamiid orders, which produce more diverse XyG structures. There is no clear correlation between the proposed diversification date and the presence of the XXGGG-type branching pattern, suggesting that this structural modification may have independently arisen in different Lamiid orders. XyGs in which <50% of the β-D-Glcp residues in the backbone are branched are also produced by various grass species (family Poaceae). These monocots are distantly related to the Asterids, again suggesting that modification of the backbone branching pattern may have occurred independently in different species. Interestingly, XyGs produced by grass species have been reported to contain no discernible amounts of α-L-Fucp residues, but to contain small amounts of l-Araf and/or d-Galp residues. Again, this suggests that modification of the branching pattern may be an adaptation that compensates for the lack of α-L-Fucp residues in the XyG. We are now determining whether the XXGGG-type branching patterns in the XyGs produced by some Lamiid species are also characteristic of the XyGs produced by various grass species. |
|
The Cellulose-Xyloglucan Network |
| The binding of cellulose is likely to be a complex topological process.
Conformational energy calculations suggest that for binding to cellulose
to occur the xyloglucan backbone must adopt a "flat ribbon" conformation
whose surface is complimentary to that of cellulose. In solution xyloglucans
are likely to adopt a "twisted" conformation that is not complementary
to cellulose. We have recently speculated that binding of xyloglucan to cellulose "untwists" the xyloglucan backbone. If both "ends" of the xyloglucan are attached to the surface this untwisting can lead to the formation of coiled structures. Duplex structures may form if the energy barrier to rotation around glycosidic bonds is sufficiently high and the anti-parallel coil is energetically stable. During wall growth XTHs may function as a polysaccharide topoisomerase that generate or relax topologically constrained structures that are formed during the assembly and restructuring of the cellulose-xyloglucan network |
 |
| Xyloglucan structure and plant growth The structure of xyloglucan has been shown to differ in a tissue specific manner. Fucosyl residues are typically absent form seed xyloglucans but present on the xyloglucans in the vegatative portions of the same plant. XLFG subunits are more abundant in pea leaf xyloglucan than in pea stem xyloglucan. We have also observed changes in the relative abundance of different subunits and the extent of their O-acetylation when suspension-cultured plant cells are grown in the presence or absence of light. We have used a monoclonal antibody (CCRC M1) that recognizes fucosylated xyloglucan to show that different cells and different parts of the same wall are differential labeled. Similary, CCRC M1 differentially labels the developing roots of the fucose deficient mur1 mutant. Only a subset of the root cells of mur1 plants produce GDP-fucose. Our observations are consistent with the notion that the extent to which xyloglucan is fucosylated in a tissue or cell is, at least in part, metabollically controlled. The role of fucosyl residues in xyloglucans is a subject of debate. Xyloglucans from solanaceous plants are not fucosylated showing that Fuc is not essential for function. Two Arabidopsis mutants in which there is a loss-of-function mutation in the xyloglycan fucosyltransferase synthesize xyloglucan that contains little (mur2) or no fucose (atfut1) grow normally under standard laboratory conditions. Nevertheless, there must be some selective pressure to maintain a viable copy of the FUT1 gene because fucose-containing side chains are present in the xyloglucans of taxonomically diverse plants (see section "Xyloglucan structure varies between plant species" above). |
| Xyloglucan-related projects Xyloglucan endoglucanase inhibitor proteins (XEGIP) We have recently discoverd that plants cells secrete a protein that binds to and inhibits fungal xyloglucan-specific endoglucanases (XEGs). We have named this Xyloglucan EndoGlucanase Inhibitor Protein (XEGIP). XEGIP has been purified, its amino acid sequence determined and a cDNA isolated. XGIP is likely to have a role in a plants defence against fungal pathogens. |
Kinetic experiments established that XEGIP inhibits XEG by forming a tightly associated 1:1 complex with a Ki of 0.5nM. No endoglucanase activity is detected when XEG is treated with a excess of XEGIP whereas mixtures containing a excess of XEG had endoglucanase activity (Figure A). The well-defined cutoff point in the inhibition of XEG by XEGIP is consistent with a 1:1 interaction stoichiometry. |
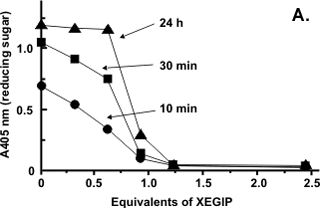 |
The association of XEGIP and XEG was demonstrated using size-exclusion chromatography (SEC) on Superdex-75 (Figure B). Free XEG (25 kDa) and XEGIP (51 kDa) are completely separated on this column. However, a component with a molecular weight greater than that of XEGIP was detected when XEG and XEGIP were mixed together and then analyzed by SEC. This higher molecular weight species was shown, using SDS-PAGE, to contain XEG and XEGIP (Qin et al., 2003). When increasing amounts of XEGIP were mixed with a fixed amount of XEG, the amount of free XEG was progressively diminished and free XEGIP was not detected in the chromatogram until XEGIP was present in molar excess over XEG (Figure B). |
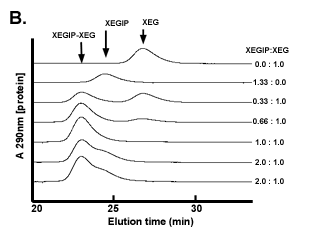 |
| Separate experiments demonstrated that XEGIP binds to immobilized XEG, that dissociation of this complex requires a strong chaotropic agent (e.g., 2 M imidazole buffer), and that the XEG retains its catalytic activity upon dissociation of the complex. Together, these results indicate that inhibition of XEG by XEGIP is a direct consequence of a 1:1 association of the two proteins. Southern analysis of genomic DNA from tomato indicated that XEGIP is present as a single-copy gene. However, five other tomato genes with 24% to 58% amino-acid sequence identity to XEGIP were identified. Genes that are homologous to XEGIP are also present in soybean, lotus, carrot, cotton, maize, rice, sorghum, Medicago trunculata, and Arabidopsis thaliana. |
XEGIPs in plant nectar In collaboration with Robert W. Thornburg at Iowa State University, we have shown that the Nectarin IV (NEC4) protein that accumulates in the nectar of ornamental tobacco plants (Nicotiana langsdorffii x Nicotiana sanderae var LxS8) (Saqlan Nakvi et al (2005) is a potent endoglucanse inhibitor. Three tryptic peptides of the protein were isolated and sequenced using tandem mass spectroscopy. These unique peptides were found to be similar to the xyloglucan-specific fungal endoglucanase inhibitor protein (XEGIP) precursor in tomato (Lycopersicon esculentum) and its homolog in potato (Solanum tuberosum). A pair of oligonucleotide primers was designed based on the potato and tomato sequences that were used to clone a 1,018-bp internal piece of nec4 cDNA from a stage 6 nectary cDNA library. The remaining portions of the cDNA were subsequently captured by 5' and 3' rapid amplification of cDNA ends. Complete sequencing of the nec4 cDNA demonstrated that NEC4 belongs to a large family of homologous proteins from a wide variety of angiosperms. Related proteins include foliage proteins and seed storage proteins. Based upon conserved identity with the wheat (Triticum aestivum) xylanase inhibitor TAXI-1, we developed a protein model showing that NEC4 contains additional amino acid loops that are not present in TAXI-1 and that glycosylation sites are surface exposed. These loops and sites of glycosylation are on the opposite face of the NEC4 molecule from the site that are believed to interact with fungal endoglycanases. Inhibition assays showed that NEC4 was a effective inhibitor (Ki of 0.35 nM) of a family GH12 xyloglucan-specific endoglucanase. No inhibitory activity was observed against family GH10 or GH11 xylanases. The patterns of expression of the NEC4 protein indicate that, while expressed in nectar at anthesis, it is most strongly expressed in the nectary gland after fertilization, indicating that inhibition of fungal cell wall-degrading enzymes may be more important after fertilization than before. |
| Enzymic synthesis of glycosides We have recently shown that oligoglycosyl (allyl, 2,3-dihydroxypropyl, ethyl, 2-hydroxyethyl, and methyl) β-glycosides are generated by endotransglycosylation reactions catalyzed by a Trichoderma reesei cellulase (York and Hawkins, 2000). This class of reactions may represent a convenient source of β-glycosides to use a synthons for the synthesis of complex glycans. |
| Tamarind xyloglucan and the immune response (In collaboration with Faith Strickland, Anderson Cancer Center, The University of Texas) Tamarind xyloglucan prevents suppresion of delayed-type hypersensitivity responses and reduced interleukin production in UV-irradiated murine epidermis. The polysaccharide also prevents suppresion of immune responses to alloantigen in mice exposed to 30kJ/m2 UVB radiation |
Glucuronoxylan in Secondary Cell Walls Mutations of Arabidopsis thaliana IRREGULAR XYLEM8 (IRX8) and IRX9 cause a collapsed xylem phenotype and decreases in xylose and cellulose in cell walls. Personn et al (2007), showed that plants carrying the irx8 mutation have reduced amounts of glucuronoxylan and homogalacturonan in their walls and suggested that IRX8 provides a link between the xylan and the secondary cell wall matrix and directly affects secondary cell wall integrity. In collaboration with Zheng-Hua Ye (Plant Biology, University of Georgia), we have characterized IRX8 and IRX9 and performed chemical and structural analyses of glucuronoxylan (GX) from irx8 and irx9 plants (Pena et al 2007). IRX8 and IRX9 are expressed specifically in cells undergoing secondary wall thickening, and their encoded proteins are targeted to the Golgi, where GX is synthesized. 1H-NMR spectroscopy showed that the reducing end of Arabidopsis GX contains the glycosyl sequence 4-β-D-Xylp-(1,4)-β-D-Xylp-(1,3)-α-L-Rhap-(1,2)-α-D-GalpA-(1,4)-D-Xylp (sequence 1), which was previously identified in birch (Betula verrucosa) and spruce (Picea abies) GX. This indicates that the reducing end structure of GXs is evolutionarily conserved in woody and herbaceous plants. This sequence is more abundant in irx9 GX than in the wild type, whereas irx8 and fragile fiber8 (fra8) plants are nearly devoid of it. The number of GX chains increased and the GX chain length decreased in irx9 plants. Conversely, the number of GX chains decreased and the chain length heterodispersity increased in irx8 and fra8 plants. Our results suggest that IRX9 is required for normal GX elongation and indicate roles for IRX8 and FRA8 in the synthesis of the glycosyl sequence at the GX reducing end. Our data, together with the results of Shimizu et al (1976) and Andersson et al (1983), show that sequence 1 is located at the reducing ends of GXs in angiosperms and gymnosperms, suggesting that evolutionarily diverse plants synthesize GX in a similar manner. Two possible biosynthetic mechanisms that differ in the direction of xylan chain elongation and the function of sequence 1 are consistent with our data. The first mechanism requires that sequence 1 is a primer for GX synthesis (A in figure) and that the backbone is extended by adding xylose to the nonreducing end. The second mechanism requires that GX is synthesized by adding monosaccharides to the reducing end (B in figure) and that transfer of sequence 1 (either one residue at a time or en bloc) to the reducing end terminates chain elongation. |
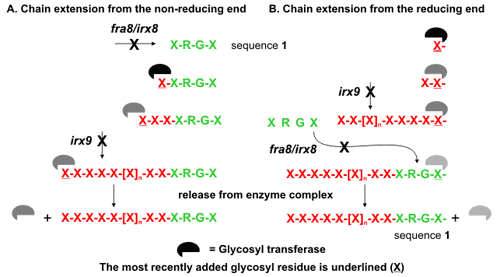 |
The synthesis of hyaluronan (Bodevin-Authelet et al., 2005; Tlapak-Simmons et al., 2005) and starch (Mukerjea and Robyt, 2005), which have been reported to occur by the addition of monosaccharides to the reducing end, serve as precedents for this mechanism. Furthermore, the presence of GX chains that do not have sequence 1 at the reducing end suggests either that this sequence is cleaved from some of the GX chains during wall biogenesis or that GX chains can be synthesized in the absence of sequence 1 as a primer. The correlation of decreased synthesis of sequence 1 with a lower proportion of GX molecules having this structure at their reducing end supports a model in which sequence 1 acts as a chain terminator. However, chain termination may still occur at relatively low frequency in the absence of sequence 1 by transfer of the nascent GX chain to water. Obtaining unambiguous evidence for the direction of xylan backbone elongation will be technically challenging, although a general approach to address this question has been proposed (Mukerjea and Robyt, 2005). Several explicit models that include specific roles for the IRX8, IRX9, and FRA8 gene products in GX biosynthesis are consistent with the available data. A common characteristic of many of these models is the requirement for multiple-subunit enzyme complexes. If this notion is correct, then it is unlikely that biochemical activity can be shown using in vitro assays based on individual gene products. The apparent complexity of the process and the products formed suggests that simple oligosaccharides may not function as acceptor substrates. Nevertheless, our observations provide a framework for the rational design of experiments to understand GX synthesis and to assign biochemical functions to specific gene products involved in this process |
|
Current Research 5. Structural characterization of xyloglucans produced by rice. 6. Structural characterizattion of glucurono- and arabino-xylans produced by diverse plants. |
|
Shimizu et al (1976). Hemicellulases of Brown Rotting Fungus, Tyromyces palustris. II. The oligosaccharides from the hydrolysate of a hardwood xylan by the intracellular xylanase. Mokuzai Gaikkashi. 22, 618-625 Andersson et al (1983). Structure of the reducing end-groups in spruce xylan. Carbohydr Res111, 283-288. York et al (1990) Structure of plant cell walls. 29. Structural analysis of xyloglucan oligosaccharides by 1H NMR spectroscopy and fast atom bombardment mass spectrometry. Carbohydr Res 200, 9-31. Hoffman et al (2005) Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae Carbohydr Res 340, 1826-1840. Jia et al (2005) NMR characterization of endogenously O-acetylated oligosaccharides isolated from tomato (Lycopersicon esculentum) xyloglucan. Carbohydr Res 340, 1818-1825. Tlapak-Simmons et al ((2005). Hyaluronan biosynthesis by class I Streptococcal hyaluronan synthases occurs at the reducing end. J Biol Chem 280, 13012-13018. Bodevin-Authelet et al (2005). Biosynthesis of hyaluronan. Direction of chain elongation. J Biol Chem 280, 8813-8818. Mukerjea and Robyt (2005). Starch biosynthessis: The primer nonreducing-end mechanism versus the nonprimer reducing-end two-site insertion mechanism. Carbohydr Res340, 245-255. Saqlan Nakvi et al (2005) Nectarin IV, a potent endoglucanase inhibitor secreted into the nectar of ornamental tobacco plants. Isolation, cloning, and characterization. Plant Physiol 139, 1389-1400. Personn et al (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which wre essential for secondary cell wall integrity. Plant Cell 19, 237-255. Pena et al (2007) Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19, 549-563. |